Comparing and Contrasting Replication, Transcription and Translation
Replication
|
Transcription
|
Translation
| |
| Initiation |
- Helicase unzips the two complementary parent strands
-Single stranded binding proteins anneal to the exposed template strands, preventing them from reannealing -Primase builds RNA primers which will be used as a starting point by DNA pol III |
-Transcription factors bind to TATA box
-RNA pol II binds to double helical DNA at the promoter region -DNA strand is unwound exposing the DNA strand - TFs, TATA box and RNA pol II, together are called the initiation complex |
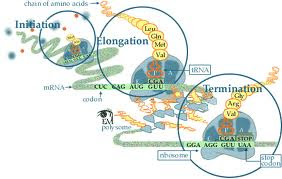
- The mRNA, a tRNA with the first amino acid and two ribosomal subunits come together.
-The ribosomal subunits assemble at the 5'cap of the mRNA transcript sandwiching the mRNA -Initiation Factors bring the large subunit such that the initiater tRNA occupies the P site. - The first tRNA brought into the P site, is carrying methionine because the start codon is AUG |
| Elongation | -DNA Pol III adds nucleotides to the free 3' end of the growing strand using template strand as a guide -leading strand, can be used by polymerases as template for a continuous complimentary strand -The lagging strand is copied away from the fork, discontinuously, in short segment known as Okazaki Fragments | RNA pol II begins building the single stranded mRNA in direction of 5' to 3', reading the DNA template in 3' to 5' direction. - The strand of DNA used for transcription is called a template strand. The strand that is not used is known as the coding strand. - This unit is known as the transcription unit. | - consists of three step cycles: - codon recognition: in this stage an elongation factor assists the hydrogen bond between the mRNA under A site with the anticodon of tRNA -Peptide bond formation: In this stage an RNA molecule catalyzes the formation of a peptide bond between the polypeptide in the P site with the amino acid in the A site. This step separates the tRNA at the P site from the growing polypeptide chain -Translocation: in this stage the ribosome moves the tRNA with the attached polypeptide from A site to the P site. Translocation ensures that mRNA is "read" 5' to 3' codon by codon |
| Termination | -DNA pol I proofreads the new strand, checking for mistakes. It also replaces the RNA primers with DNAnucleotides -DNA Ligase then "glues" the gaps between the Okazaki fragments. | -The mRNA is synthesized till RNA polymerase recognizes the termination sequence at the end of a gene, known as the terminator sequence (AAUAAA) Posttranscriptional Modifications (in eukaryotic cells): - 5' cap is added to the start of the primary transcript -Poly A tail is added to the 3' end - Introns (non-coding parts of the DNA), are taken out by proteins known as spliceosomes ---> Once the primary transcript has been capped and tailed and the intros excised, the process transcript is known as mRNA(messengerRNA) We are now ready to send the mRNA out into the cytoplasm | -Termination occurs when one of the stop codons: UAG UGA UAA reaches the A site - A release factor binds to the stop codon and hydrolyzes the bond between the polypeptide and its tRNA in the P site - Now the polypeptide chain is free and the translation complex disassembles. |
 |
Replication |
 |
| Translation |





